Sanger Method of DNA Sequencing: Last week, we explained “What is the Maxam Gilbert Method of DNA Sequencing?” and many readers reached out saying they wanted to know how the Sanger method of DNA sequencing works. So, here we are!
DNA sequencing is simply the process of determining the exact order of nucleotides, adenine (A), thymine (T), cytosine (C), and guanine (G), in a DNA strand. While several techniques have evolved over the years, the Sanger dideoxy method of DNA sequencing remains a foundation stone. In fact, despite the rise of next-generation sequencing (NGS), scientists still rely on the Sanger approach for validation, small DNA fragments, and clinical applications.
In this blog, we will explore the Sanger method of DNA sequencing, one of the most widely used and historically significant techniques in molecular biology. We’ll begin by understanding what the Sanger method is and the principle behind how it and learn about the applications of the Sanger method in research and diagnostics.
As Frederick Sanger, the man behind this technique, once remarked, “DNA sequencing is like reading the book of life, letter by letter.”
Read More: Let’s Talk DNA: What is the Maxam Gilbert Method of DNA Sequencing?
What is the Sanger Method of DNA Sequencing?
The Sanger method of DNA sequencing, or the Sanger method, or dideoxy sequencing is also called the dideoxy chain termination method. It was developed in 1977 by Frederick Sanger and colleagues. At its core, the method uses special building blocks called dideoxynucleotides (ddNTPs) that stop DNA synthesis whenever they’re added to a growing chain.
So, instead of getting one long DNA product, you get many fragments of different lengths, each ending at a specific nucleotide. When you separate these fragments and read them, you can reconstruct the full sequence of the DNA.
The Sanger dideoxy method of DNA sequencing was the gold standard for decades. It played a central role in the Human Genome Project, which decoded the entire human genome. Even today, when someone asks “what is the Sanger method of DNA sequencing?”, the simplest answer is: it’s a method to read DNA sequences using chain-terminating nucleotides.
Principle of Sanger’s Method of DNA Sequencing
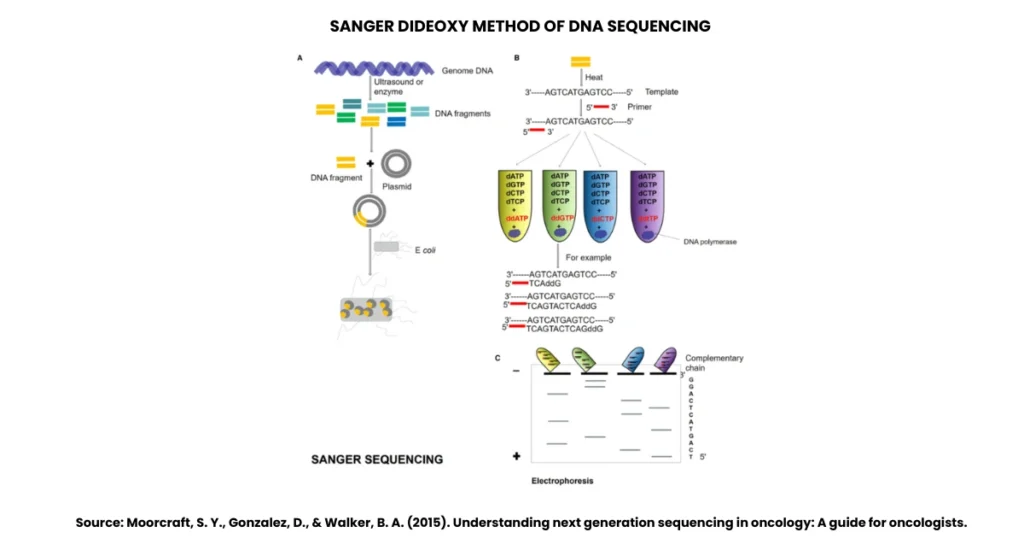
The principle of Sanger’s method of DNA sequencing lies in controlled interruption of DNA synthesis. Here’s the breakdown:
- DNA synthesis requires four normal nucleotides (dNTPs).
- If you add a special nucleotide, dideoxynucleotide (ddNTP), the synthesis stops. Why? Because ddNTPs lack a hydroxyl group (-OH) at the 3’ position of the sugar. Without this -OH, the next nucleotide can’t attach, leading to chain termination.
- By running four parallel reactions (A, T, C, G), each with one type of ddNTP mixed with normal nucleotides, you generate fragments ending at each possible position of that base.
- Separating these fragments and reading them in order gives the DNA sequence.
In simple words, normal nucleotides build the chain, while ddNTPs act as full stops.
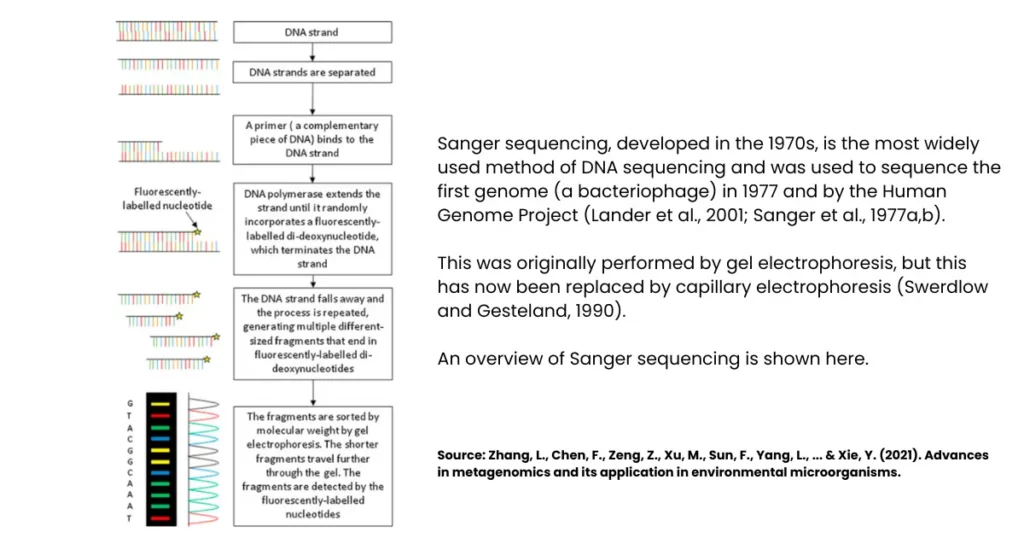
Steps of Sanger Dideoxy Method of DNA Sequencing
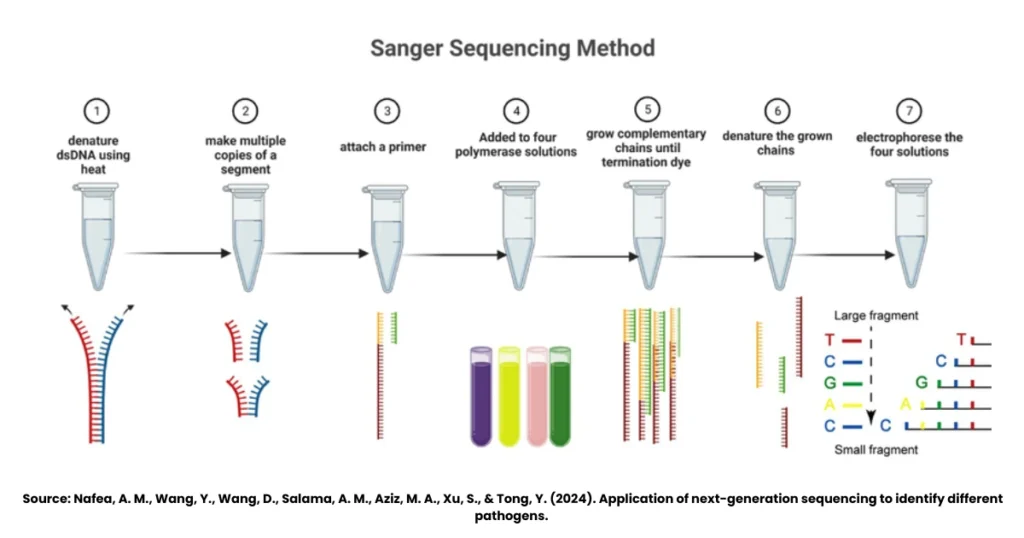
The Sanger dideoxy method of DNA sequencing follows a clear sequence of laboratory steps. Let’s decode them one by one:
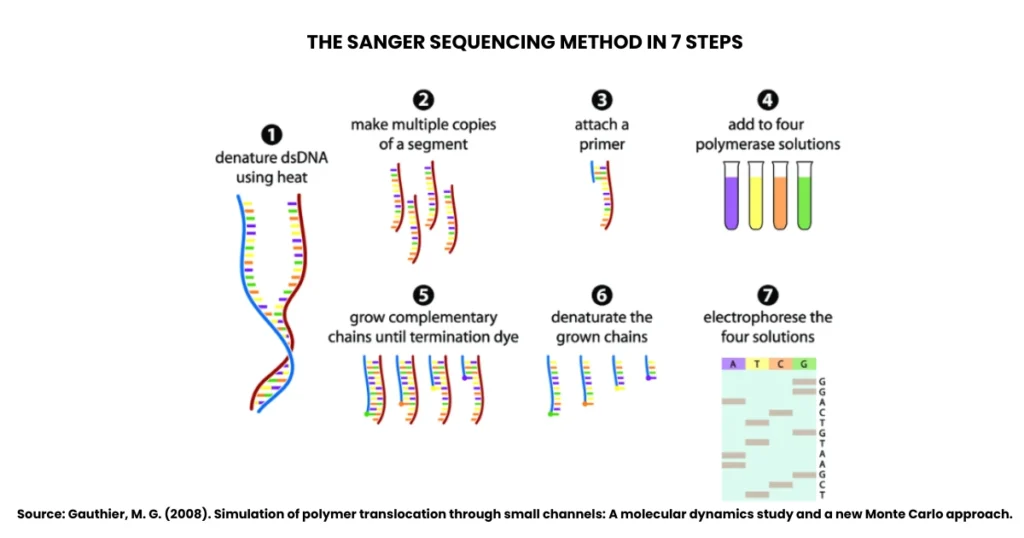
1. Denature Double-Stranded DNA Using Heat
The process begins with the double-stranded DNA (dsDNA) that needs to be sequenced. Heat is applied to denature or “unzip” the two strands. This makes the DNA single-stranded and ready for sequencing.
2. Make Multiple Copies of a DNA Segment
The single-stranded DNA is then copied to create multiple identical fragments. This step ensures that there’s enough DNA material to work with, because sequencing requires thousands of copies for accurate detection.
3. Attach a Primer
A short primer is added to the single-stranded DNA. This primer binds to the template and gives DNA polymerase a starting point to begin the synthesis. Without this primer, the reaction wouldn’t proceed.
4. Add to Four Polymerase Solutions
The DNA template with primer is divided into four separate reaction tubes. Each tube contains:
- DNA polymerase enzyme
- All four standard nucleotides (dNTPs)
- A small amount of one dideoxynucleotide (ddNTP), labelled with a fluorescent dye (ddATP, ddTTP, ddCTP, or ddGTP).
Each tube therefore has a different ddNTP.
5. Grow Complementary Chains Until Termination by Dye
DNA polymerase starts synthesising new DNA strands by adding nucleotides. However, whenever a ddNTP is incorporated instead of a normal nucleotide, the chain terminates. This produces DNA fragments of varying lengths, each ending with a labelled ddNTP.
6. Denature the Grown Chains
Once the complementary DNA chains are formed, they are denatured again to separate them into single strands. This step is necessary before separating them by size.
7. Electrophorese the Four Solutions
Finally, the DNA fragments from each reaction are separated using gel electrophoresis or, more commonly today, capillary electrophoresis. The fluorescent tags on ddNTPs help detect which nucleotide is present at the end of each fragment. The order of fragments reveals the full DNA sequence.
Sanger Method of DNA Sequencing Flow Chart
This Sanger method of DNA sequencing flow chart makes it clear: start with template -> terminate with ddNTPs -> separate -> read sequence.Application of Sanger Method of DNA Sequencing
The application of Sanger method of DNA sequencing is vast and still relevant despite the boom of NGS. Some key uses of it are as follows:
- Clinical diagnostics: Detecting mutations in genes like BRCA1/2 linked to breast cancer.
- Forensic science: Identifying individuals from DNA evidence.
- Molecular biology: Confirming cloned DNA sequences before further experiments.
- Microbiology: Sequencing bacterial genes for strain identification.
- Evolutionary biology: Comparing DNA sequences of different organisms.
Advantages and Disadvantages of Sanger Method
Every technique has its pros and cons and right now, let’s explore the advantages and disadvantages of Sanger method in detail.
Advantages
- High accuracy: Error rate is extremely low (~0.001%).
- Read length: Can sequence up to 900 base pairs in one run.
- Cost-effective for small projects: Ideal for single genes or small DNA fragments.
- Reliable validation tool: Often used to confirm results obtained from NGS.
Disadvantages
- Low throughput: Not suitable for large genomes due to time and cost.
- Expensive for big projects: Sequencing an entire genome is impractical with Sanger.
- Labour-intensive: Requires multiple reactions and manual handling compared to automated NGS platforms.
Thus, while the advantages and disadvantages of Sanger method highlight its limitations, the technique remains powerful for targeted sequencing.
Comparison: Sanger vs Maxam-Gilbert vs NGS
- Sanger vs Maxam-Gilbert: Sanger is safer (no toxic chemicals), easier, and more widely adopted.
- Sanger vs NGS: NGS is faster and cheaper for whole genomes, but Sanger is still unmatched in accuracy for small fragments.
This makes the Sanger dideoxy method of DNA sequencing not obsolete but complementary to modern tools.
On A Final Note…
The Sanger method of DNA sequencing is a timeless tool in genetics. From its principle of using chain-terminating ddNTPs to its workflow illustrated in a Sanger method of DNA sequencing flow chart, the method is still widely taught and practiced. Its reliability ensures that even in the age of Next-generation sequencing (NGS), it holds a strong place!
If you’re a student, researcher, or just curious about biotechnology, understanding the application of Sanger method of DNA sequencing, and weighing the advantages and disadvantages of Sanger method, gives you a strong foundation to appreciate how far we’ve come in decoding life’s blueprint.
As Sanger’s work reminds us: “Progress in science depends on new techniques, new discoveries, and new ideas, probably in that order.”
FAQs
Q1. What is the Sanger method of DNA sequencing in simple words?
It’s a DNA reading technique where chain-terminating nucleotides stop DNA synthesis, creating fragments that can be read to determine the sequence.
Q2. What is the principle of Sanger’s method of DNA sequencing?
It’s based on selective incorporation of dideoxynucleotides (ddNTPs) that terminate chain elongation.
Q3. What is the Sanger method of DNA sequencing flow chart?
A step-by-step guide: setup, synthesis, chain termination, separation and reading sequence.
Q4. What is an application of Sanger method of DNA sequencing?
It’s used in medical diagnostics, confirming cloned DNA, microbial identification, and forensics.
Q5. What are the advantages and disadvantages of Sanger method?
Advantages are accurate, long reads, reliable. Disadvantages are that it is costly and slow for large genomes.

